Permutation-based selection in the melanoma data
[1]:
import os
import pandas as pd
import numpy as np
import anndata as ann
import spatialdm as sdm
from spatialdm.datasets import dataset
import spatialdm.plottings as pl
import matplotlib.pyplot as plt
print("SpatailDM version: %s" %sdm.__version__)
SpatailDM version: 0.0.7
The melanoma dataset from Thrane, et al. was publicly available, and we obtained raw counts, pre-processed log counts, and spatial coordinates from Matt Stone (Git repository). For easier reuse, we included them in an anndata object which can be loaded directly in SpatialDM Python package.
[3]:
adata = dataset.melanoma()
/home/yoyo/miniconda2/envs/CC/lib/python3.9/site-packages/anndata/_core/anndata.py:121: ImplicitModificationWarning: Transforming to str index.
warnings.warn("Transforming to str index.", ImplicitModificationWarning)
adata.X,adata.raw,adata.obs,adata.obsm['spatial']SpatialDM object and preprocessing
We generate a suitable weight matrix for the SpatialDM object at first. Here considering the scale of the spatial coordinates and spot-spot distance (200 micrometers here), we set radial basis kernel parameter l = 1.2, and trimmed all weights < 0.2 (cutoff) to match the normal range of CCC (200 micrometers, 1 spot away from the sender cell here)
Note Alternative to the cutoff parameter, we can set n_neighbors to around 8 to restrain the range of CCC.
[5]:
sdm.weight_matrix(adata, l=1.2, cutoff=0.2, single_cell=False) # weight_matrix by rbf kernel
/home/yoyo/miniconda2/envs/CC/lib/python3.9/site-packages/sklearn/utils/validation.py:70: FutureWarning: Pass n_neighbors=6 as keyword args. From version 1.0 (renaming of 0.25) passing these as positional arguments will result in an error
warnings.warn(f"Pass {args_msg} as keyword args. From version "
Note
If the scale of the spatial coordinates is larger (e.g. several thousand in the intestinal example) or smaller, or when the spot-spot distance varies (which is common for different platforms), the l can be very different. It’s a crucial step to determine a suitable l to match the biological context. We recommend the following plotting to check the resulting weight matrix from the previous step. If needed, users can run weight_matrix iteratively to decide the most optimal l and
cutoff.
[6]:
# visualize range of interaction
import matplotlib.pyplot as plt
plt.figure(figsize=(3,3))
# plt.scatter(adata.obsm['spatial'][:,0], adata.obsm['spatial'][:,1],
# c=adata.obsp['weight'][50])
plt.scatter(adata.obsm['spatial'][:,0], adata.obsm['spatial'][:,1],
c=adata.obsp['weight'].A[50])
[6]:
<matplotlib.collections.PathCollection at 0x7fd4b64386d0>
Next step is to extract valid LR pairs from the database (by default, we use CellChatDB). Filtering out sparsely expressed ligand or receptor (e.g. 3) can help us get more interpretable results later.
[7]:
sdm.extract_lr(adata, 'human', min_cell=3) # find overlapping LRs from CellChatDB
Global and local selection (permutation approach)
It is a crucial step to identify dataset-specific interacting LR pairs (global selection) for ensuring quality analysis and reliable interpretation of the putative CCC. With high computation efficiency, SpatialDM can complete global selection in seconds.
[13]:
import time
start = time.time()
sdm.spatialdm_global(adata, 1000, specified_ind=None, method='both', nproc=1) # global Moran selection
sdm.sig_pairs(adata, method='permutation', fdr=True, threshold=0.1) # select significant pairs
print("%.3f seconds" %(time.time()-start))
100%|██████████| 1000/1000 [00:06<00:00, 155.27it/s]
8.346 seconds
We used fdr corrected global p-values and a threshold FDR < 0.1 (default) to determine which pairs to be included in the following local identification steps. There are 103 pairs being selected in this data.
[14]:
print(adata.uns['global_res'].selected.sum())
adata.uns['global_res'].sort_values(by='fdr').head()
103
[14]:
| Ligand0 | Ligand1 | Ligand2 | Receptor0 | Receptor1 | Receptor2 | z_pval | z | perm_pval | fdr | selected | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CSPG4_ITGA2_ITGB1 | CSPG4 | None | None | ITGA3 | ITGB1 | None | 1.372040e-06 | 4.689101 | 0.0 | 0.0 | True |
| PECAM1_PECAM1 | PECAM1 | None | None | PECAM1 | None | None | 2.792191e-52 | 15.170039 | 0.0 | 0.0 | True |
| SEMA3D_NRP2_PLXNA1 | SEMA3D | None | None | NRP2 | PLXNA1 | None | 1.267594e-04 | 3.658679 | 0.0 | 0.0 | True |
| HLA-C_CD8A | HLA-C | None | None | CD8A | None | None | 7.035725e-07 | 4.823990 | 0.0 | 0.0 | True |
| COL6A1_ITGA1_ITGB1 | COL6A1 | None | None | ITGA1 | ITGB1 | None | 8.586635e-06 | 4.298790 | 0.0 | 0.0 | True |
Local selection is then carried out for the selected 103 pairs to identify where the LRI take place, in a single-spot resolution.
[15]:
start = time.time()
sdm.spatialdm_local(adata, n_perm=1000, method='both', specified_ind=None, nproc=1) # local spot selection
sdm.sig_spots(adata, method='permutation', fdr=False, threshold=0.1) # significant local spots
print("%.3f seconds" %(time.time()-start))
/home/yoyo/miniconda2/envs/CC/lib/python3.9/site-packages/anndata/_core/raw.py:139: FutureWarning: X.dtype being converted to np.float32 from int64. In the next version of anndata (0.9) conversion will not be automatic. Pass dtype explicitly to avoid this warning. Pass `AnnData(X, dtype=X.dtype, ...)` to get the future behavour.
return anndata.AnnData(
100%|██████████| 1000/1000 [00:00<00:00, 3934.37it/s]
100%|██████████| 1000/1000 [00:00<00:00, 4645.87it/s]
1.115 seconds
By default, local selection is performed for all selected pairs in the previous step.
Note
Apply an array of integer indices of selected pairs to select_num to run local selection in selected pairs
The global and local results are easily accessible through global_res and local_perm_p or local_z_p
Visualize pair(s)
SpatialDM provides plotting utilities for general summary of global results:
[17]:
pl.global_plot(adata, pairs=['SPP1_CD44', 'CSF1_CSF1R', 'VEGFA_VEGFR2', 'ANGPTL4_SDC2'],
figsize=(6,5), cmap='RdGy_r', vmin=-1.5, vmax=2)

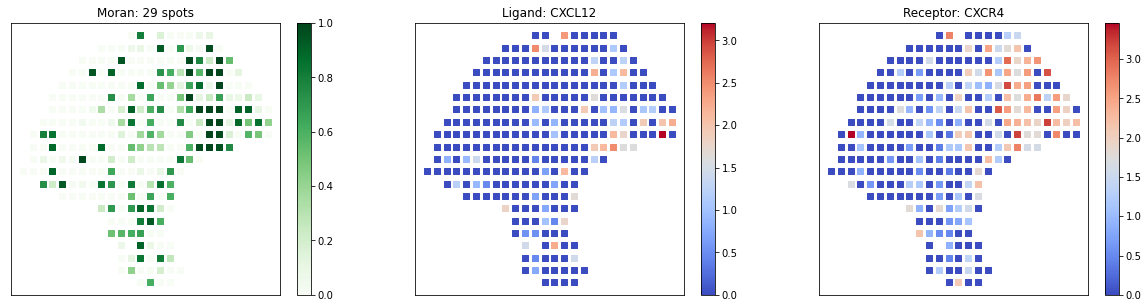
Known melanoma-related pairs were all observed in the selected pairs. It would then be meaningful to investigate the spatial context of their interactions.
[18]:
# visualize known melanoma pairs
pl.plot_pairs(adata, ['CSF1_CSF1R', 'VEGFA_VEGFR2', 'CXCL12_CXCR4'], marker='s')
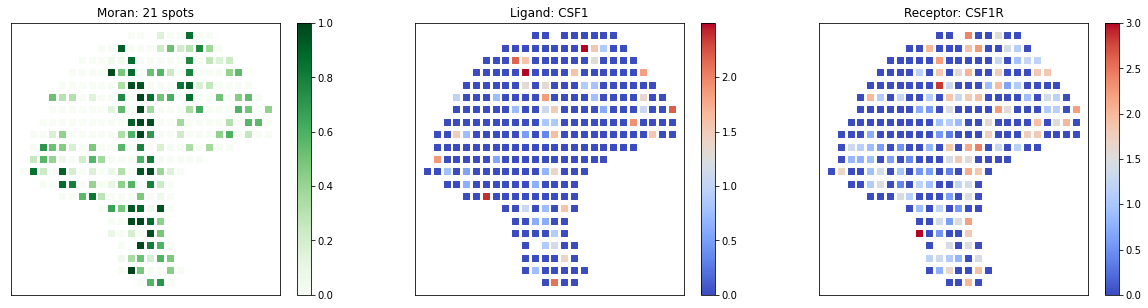
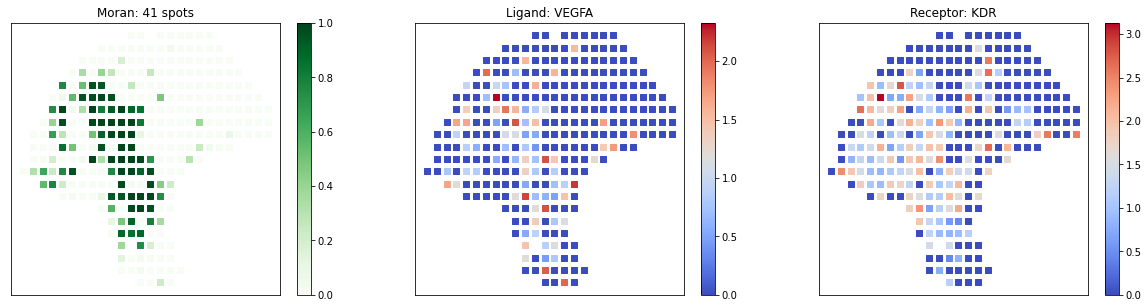
Spatial Clustering of Local Spots
SpatialDE allows clustering of spatially auto-correlated genes. Here, we repurposed SpatialDE to identify spatially auto-correlated interactions by using binary local selection status as input.
[19]:
import NaiveDE
import SpatialDE
Filter out sparse interactions with fewer than 3 identified interacting spots. Cluster into 6 patterns.
[20]:
# SpatialDE code
bin_spots = adata.uns['selected_spots'].astype(int)[adata.uns['local_stat']['n_spots']>2]
print(bin_spots.shape[0], " pairs used for spatial clustering")
103 pairs used for spatial clustering
[21]:
from threadpoolctl import threadpool_limits
with threadpool_limits(limits=1, user_api='blas'):
results = SpatialDE.run(adata.obsm['spatial'], bin_spots.transpose())
histology_results, patterns = SpatialDE.aeh.spatial_patterns(
adata.obsm['spatial'], bin_spots.transpose(), results, C=3, l=3, verbosity=1)
iter 0, ELBO: -2.99e+10
iter 1, ELBO: -1.57e+10, delta_ELBO: 1.42e+10
iter 2, ELBO: -1.57e+10, delta_ELBO: 4.65e+03
iter 3, ELBO: -1.57e+10, delta_ELBO: 1.29e+03
iter 4, ELBO: -1.57e+10, delta_ELBO: 3.45e+00
iter 5, ELBO: -1.57e+10, delta_ELBO: 4.57e+00
iter 6, ELBO: -1.57e+10, delta_ELBO: 1.31e+00
iter 7, ELBO: -1.57e+10, delta_ELBO: 1.17e+00
iter 8, ELBO: -1.57e+10, delta_ELBO: 3.26e+01
iter 9, ELBO: -1.57e+10, delta_ELBO: 2.56e+01
iter 10, ELBO: -1.57e+10, delta_ELBO: 2.93e-01
iter 11, ELBO: -1.57e+10, delta_ELBO: 4.65e-04
Converged on iter 11
[22]:
plt.figure(figsize=(9,8))
for i in range(3):
plt.subplot(2, 2, i + 2)
plt.scatter(adata.obsm['spatial'][:,0], adata.obsm['spatial'][:,1], marker = 's',c=patterns[i], s=35);
plt.axis('equal')
pl.plt_util('Pattern {} - {} genes'.format(i, histology_results.query('pattern == @i').shape[0] ))
plt.savefig('mel_DE_clusters.pdf')
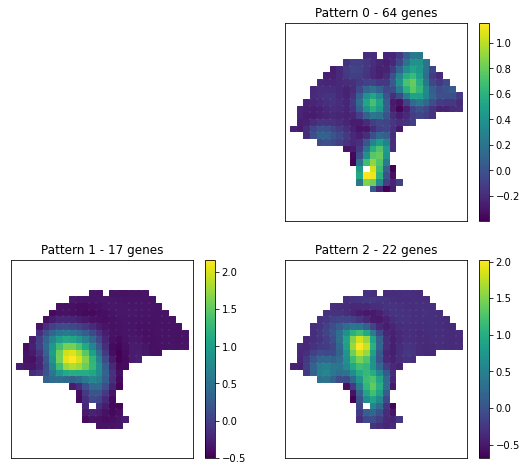
Note For detail parameter settings like C and l, please refer to SpatialDE tutorial
SpatialDM provides compute_pathway function to group a list of interactions based on the pathways they belong. The input can be a dictionary of several lists.
[23]:
dic=dict()
for i in histology_results.sort_values('pattern').pattern.unique():
dic['Pattern_{}'.format(i)]=histology_results.query('pattern == @i').sort_values('membership')['g'].values
Save the anndata file to retrieve later
[25]:
adata_out = adata.copy()
adata_out.uns['local_stat']['local_permI'] = 'None'
adata_out.uns['local_stat']['local_permI_R'] = 'None'
sdm.write_spatialdm_h5ad(adata_out, filename='mel_sdm_out.h5ad')
# sdm.read_spatialdm_h5ad(filename='mel_sdm_out.h5ad')
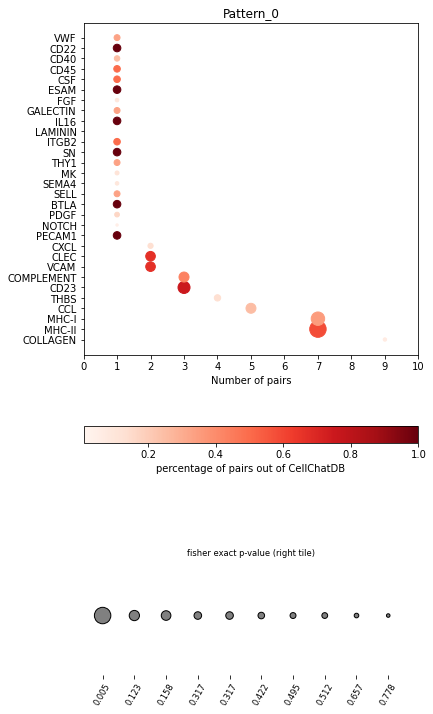
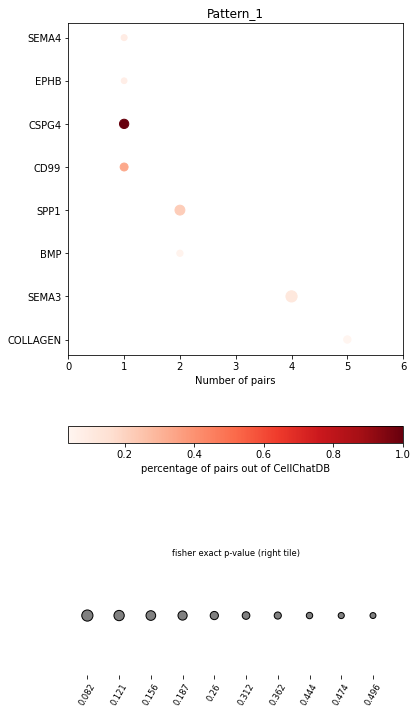
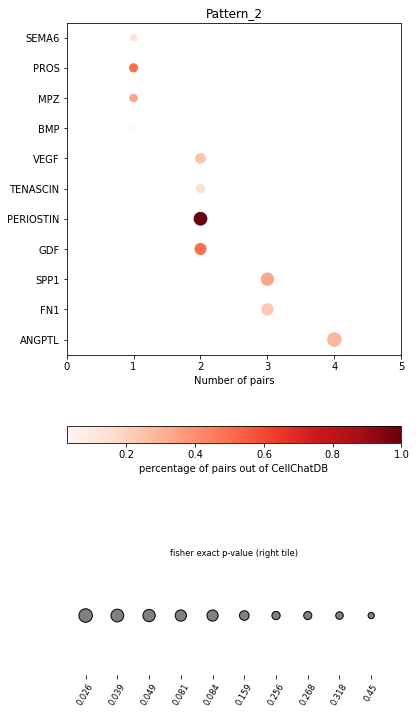
In the dot plot of the lymphoid-associated pattern, we identified CD23 and other immune-related pathways.
[28]:
pl.dot_path(adata, dic=dic, figsize=(6,12))
SpatialDM provides chord_celltype and other chord diagrams to visualize the aggregated cell types (or interactions)
[30]:
adata.obsm['celltypes'] = adata.obs[adata.obs.columns]
pl.chord_celltype(adata, pairs=['FCER2A_CR2'], ncol=1, min_quantile=0.01)
# save='mel_FCER2A_CR2_ct.svg'
[30]:
alternative DE input with Moran R values
[16]:
# local_sum = adata.uns['selected_spots'].copy()
# local_sum=local_sum.transpose()
# local_sum[local_sum.columns] =(adata.uns['local_stat']['local_I'] + adata.uns['local_stat']['local_I'])
# results = SpatialDE.run(adata.obsm['spatial'], local_sum)
# histologyesults, patterns = SpatialDE.aeh.spatial_patterns(adata.obsm['spatial'], local_sum,
# results, C=3, l=5,
# verbosity=1)
# plt.figure(figsize=(9,8))
# for i in range(3):
# plt.subplot(2, 2, i + 1)
# plt.scatter(adata.obsm['spatial'][:,0], adata.obsm['spatial'][:,1], marker = 's',c=patterns[i], s=35);
# plt.axis('equal')
# pl.plt_util('Pattern {} - {} genes'.format(i, histologyesults.query('pattern == @i').shape[0] ))
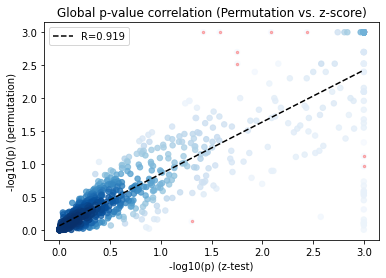
Consistency between permutation and z-score approaches
As an alternative to permutation method, we derived the first and second moments of the null distribution to analytically obtain a z-score and its according p-values, which are highly consistent with the permutation method.
[31]:
# global consistency
x = -np.log10(adata.uns['global_res'].z_pval.values)
y = -np.log10(adata.uns['global_res'].perm_pval)
x = np.where(x>3, 3, x)
y = np.where(np.isinf(y), 3, y)
pl.corr_plot(x, y, method='spearman')
plt.xlabel('-log10(p) (z-test)')
plt.ylabel('-log10(p) (permutation)')
plt.title('Global p-value correlation (Permutation vs. z-score)')
plt.savefig('mel_perm_z_corr.pdf')
/home/yoyo/miniconda2/envs/CC/lib/python3.9/site-packages/pandas/core/arraylike.py:364: RuntimeWarning: divide by zero encountered in log10
result = getattr(ufunc, method)(*inputs, **kwargs)
[32]:
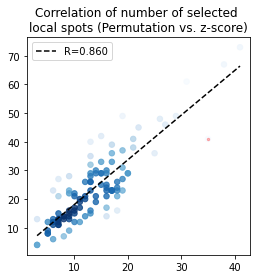
# local consistency
plt.figure(figsize=(4,4))
x=(adata.uns['local_z_p']<0.1).sum(1).values
y=(adata.uns['local_perm_p']<0.1).sum(1).values
pl.corr_plot(x, y, method='pearson')
plt.title('Correlation of number of selected \nlocal spots (Permutation vs. z-score)')
plt.savefig('mel_local_cor.pdf')
Cell type prediction with local Moran
[34]:
from sklearn.linear_model import LinearRegression
Note The prediction results may differ by different annotation accuracy and potentially other factor. Here we generated the cell type decomposition results using RCTD.
[35]:
X = adata.uns['local_perm_p'].values
y=adata.obs.values
reg = LinearRegression().fit(X.T, y)
y_pred = reg.predict(X.T)
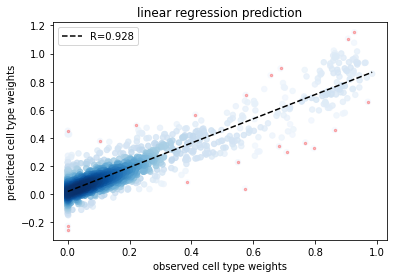
pl.corr_plot(np.hstack(y),np.hstack(y_pred), method='pearson')
plt.xlabel('observed cell type weights')
plt.ylabel('predicted cell type weights')
plt.title('linear regression prediction')
# plt.savefig('linear_regression_celltype.pdf')
[35]:
Text(0.5, 1.0, 'linear regression prediction')